Siena Galaxy Atlas 2020
Welcome to the data portal for the SGA-2020!
Please refer to the SGA-2020 documentation (or click on the SGA dropdown menu) for full details regarding this data release and the required acknowledgments and conditions of use.
We also encourage you to read the paper!
Full Catalog [675 MB]
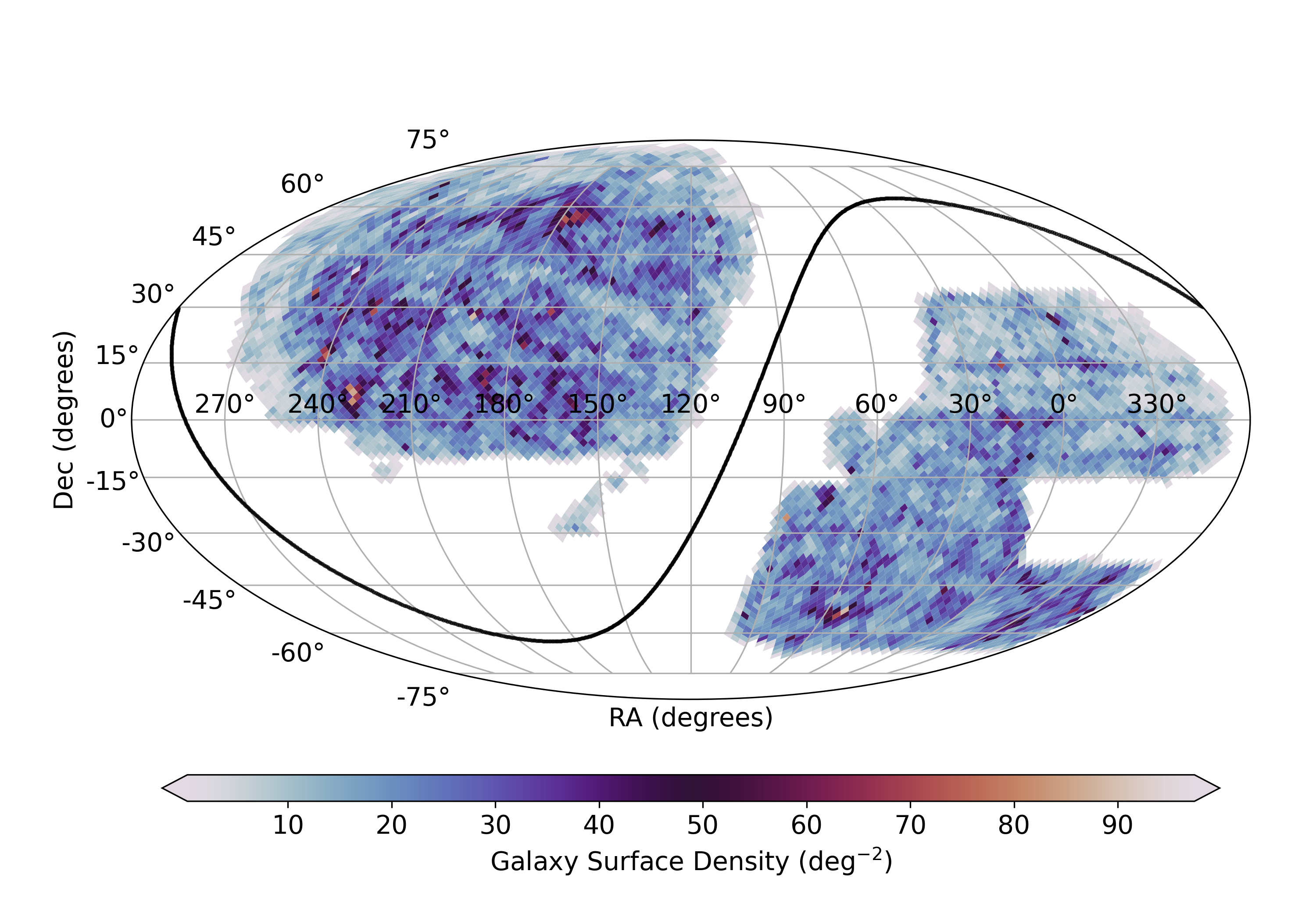
SGA-2020.fits (data model)On the web
https://portal.nersc.gov/project/cosmo/data/sga/2020/dataAt NERSC (for collaborators)
/global/cfs/cdirs/cosmo/data/sga/2020Simple Filter
Cone Search
| Number | SGA ID | Group Name | Galaxy | RA (degrees) |
Dec (degrees) |
Galaxy Diameter (arcmin) |
|
|---|---|---|---|---|---|---|---|
 |
1 | 495237 | PGC1961515 | PGC1961515 | 0.0004660 | 31.731567 | 0.692 |
 |
2 | 942428 | PGC520795 | PGC520795 | 0.0006385 | -45.954255 | 0.685 |
 |
3 | 1431578 | PGC535833 | PGC535833 | 0.0006907 | -44.578090 | 0.374 |
 |
4 | 435797 | PGC124374 | PGC124374 | 0.0011596 | -41.422890 | 0.669 |
 |
5 | 859355 | PGC1985872 | PGC1985872 | 0.0023610 | 32.209246 | 1.169 |
 |
6 | 492070 | PGC143101 | PGC143101 | 0.0025998 | -59.012969 | 0.594 |
 |
7 | 265575 | PGC582652 | PGC582652 | 0.0030718 | -40.770465 | 0.504 |
 |
8 | 1433046 | PGC1283524 | PGC1283524 | 0.0030873 | 5.442523 | 0.781 |
 |
9 | 171414 | PGC402770 | PGC402770 | 0.0033439 | -56.140738 | 0.616 |
 |
10 | 944480 | PGC1205857 | PGC1205857 | 0.0035290 | 1.787322 | 0.805 |
 |
11 | 400920 | PGC130936 | PGC130936 | 0.0036306 | -40.903238 | 1.441 |
 |
12 | 1409432 | PGC2009149 | PGC2009149 | 0.0050164 | 32.710246 | 0.758 |
 |
13 | 381301 | PGC1627451 | PGC1627451 | 0.0051356 | 20.442654 | 0.635 |
 |
14 | 524063 | PGC2025207 | PGC2025207 | 0.0065838 | 33.165002 | 0.481 |
 |
15 | 243047 | PGC427855 | PGC427855 | 0.0066706 | -53.631874 | 0.430 |
 |
16 | 515342 | PGC1004399 | PGC1004399 | 0.0068807 | -8.385656 | 0.640 |
 |
17 | 418594 | PGC597378 | PGC597378 | 0.0073157 | -39.512843 | 0.386 |
 |
18 | 462872 | SDSSJ000001.99+154149.8 | SDSSJ000001.99+154149.8 | 0.0083455 | 15.697169 | 0.570 |
 |
19 | 848636 | PGC000006 | PGC000006 | 0.0089514 | 15.881594 | 0.604 |
 |
20 | 405917 | PGC1032184 | PGC1032184 | 0.0089642 | -6.496401 | 0.494 |
 |
21 | 668284 | PGC1281052 | PGC1281052 | 0.0095881 | 5.288252 | 0.810 |
 |
22 | 950344 | PGC371047 | PGC371047 | 0.0116891 | -59.489296 | 0.714 |
 |
23 | 433498 | PGC923433 | PGC923433 | 0.0121281 | -14.529781 | 0.642 |
 |
24 | 952036 | PGC143105 | PGC143105 | 0.0128957 | -34.064014 | 1.017 |
 |
25 | 322842 | PGC1308497 | PGC1308497 | 0.0131870 | 6.838189 | 0.502 |
 |
26 | 822461 | PGC974615 | PGC974615 | 0.0138724 | -10.721079 | 1.083 |
 |
27 | 777969 | PGC000004 | PGC000004 | 0.0142396 | 23.087555 | 1.390 |
 |
28 | 1239395 | PGC1964345 | PGC1964345 | 0.0147152 | 31.785639 | 0.964 |
 |
29 | 860508 | PGC073199 | PGC073199 | 0.0150749 | 26.013679 | 1.028 |
 |
30 | 524198 | PGC129171 | PGC129171 | 0.0169975 | -49.281085 | 0.977 |
 |
31 | 1299668 | PGC445008 | PGC445008 | 0.0175278 | -52.365842 | 0.797 |
 |
32 | 101600 | PGC000007 | PGC000007 | 0.0184946 | -0.083366 | 1.021 |
 |
33 | 1172660 | PGC997239 | PGC997239 | 0.0192229 | -8.943797 | 0.473 |
 |
34 | 532980 | PGC1045312 | PGC1045312 | 0.0197103 | -5.392681 | 0.493 |
 |
35 | 1261713 | PGC143106 | PGC143106 | 0.0197839 | 0.781712 | 0.664 |
 |
36 | 1297926 | PGC000015 | PGC000015 | 0.0211679 | -5.209407 | 0.819 |
 |
37 | 851586 | PGC1815123 | PGC1815123 | 0.0216969 | 27.683065 | 1.188 |
 |
38 | 587786 | PGC1344920 | PGC1344920 | 0.0224463 | 8.343027 | 0.653 |
 |
39 | 1214984 | PGC1829164_GROUP | PGC1828876 | 0.0229193 | 28.191468 | 0.723 |
 |
40 | 1346323 | PGC674186 | PGC674186 | 0.0234048 | -33.596269 | 0.440 |
 |
41 | 780957 | PGC407967 | PGC407967 | 0.0244105 | -55.578330 | 0.561 |
 |
42 | 165751 | PGC199318 | PGC199318 | 0.0246017 | -35.657913 | 0.695 |
 |
43 | 486522 | PGC1484403 | PGC1484403 | 0.0248112 | 15.429921 | 0.604 |
 |
44 | 248739 | PGC1829164_GROUP | PGC1829164 | 0.0252431 | 28.202046 | 0.969 |
 |
45 | 1134221 | PGC143109 | PGC143109 | 0.0279933 | -7.875016 | 0.799 |
 |
46 | 655345 | PGC1814613 | PGC1814613 | 0.0291227 | 27.664545 | 0.483 |
 |
47 | 675492 | UGC12890 | UGC12890 | 0.0293128 | 8.279183 | 2.911 |
 |
48 | 960214 | PGC311006 | PGC311006 | 0.0295792 | -65.695669 | 0.678 |
 |
49 | 1074239 | PGC1561449 | PGC1561449 | 0.0296227 | 18.503455 | 0.695 |
 |
50 | 1163974 | PGC357331 | PGC357331 | 0.0297368 | -61.184925 | 0.672 |